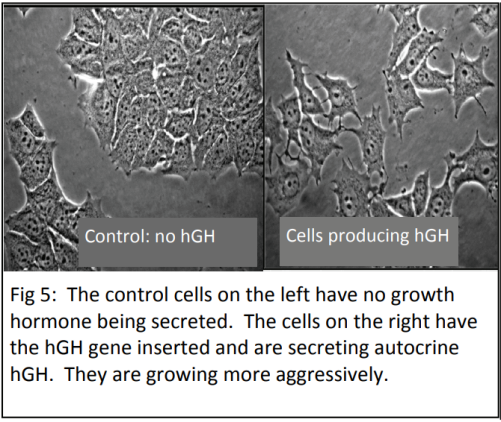
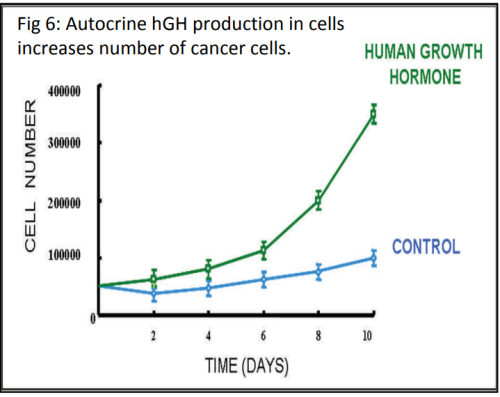
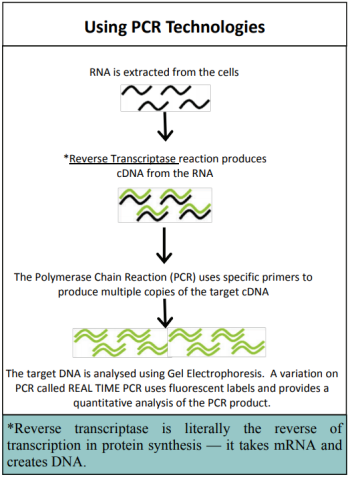
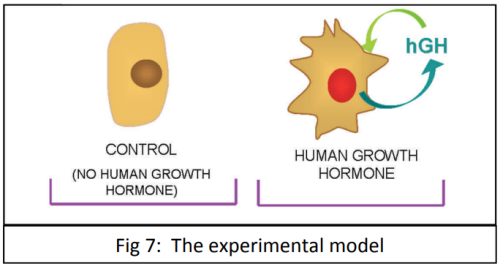
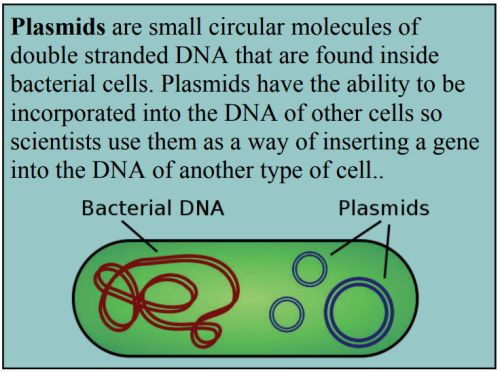
When studying disease, scientists want to find out how gene expression is being affected in the cells (e.g. genes being switched on/off or increasing/decreasing the level of gene expression). The phenotypic response that we see in organisms, such as the development of breast cancer, is almost always the result of a group of genes being expressed together, rather than just one gene. Scientists have been able to study gene expression one gene at a time for many years using a biotechnological process called the polymerase chain reaction (PCR). PCR is used to amplify (make multiple copies of) DNA.
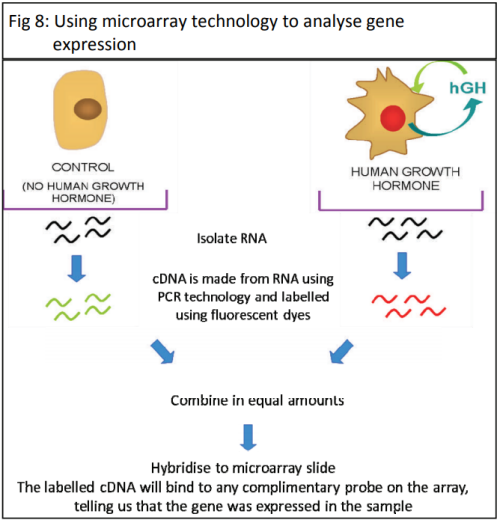
In gene expression studies, PCR is used to amplify specific genes to find out whether that gene is being expressed in the tissue being studied. If genes are being expressed then mRNA will be produced. This mRNA is extracted from the cells and a process called reverse transcriptase is used to make a short section of DNA that is complementary to the mRNA - called cDNA. The cDNA is slightly different from the original DNA because it does not contain introns (a segment of a DNA or RNA molecule which does not code for proteins and interrupts the sequence of genes). The amount of cDNA produced will depend on the amount of mRNA which is determined by how active the gene is.
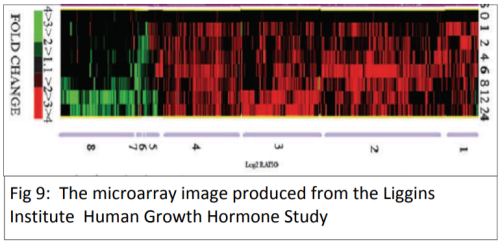
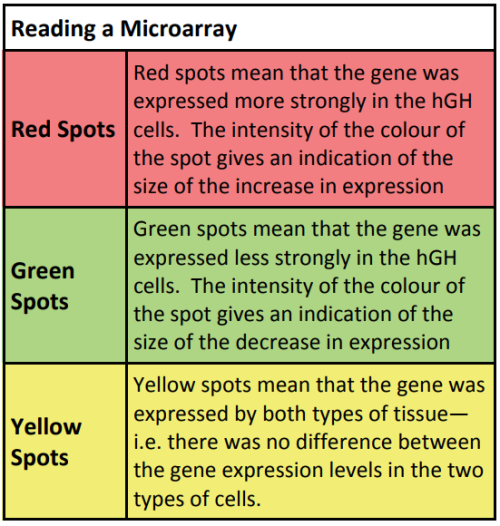
The cDNA is then amplified and analysed to establish how much mRNA was in the original sample. This gives an indication of how active the gene was. PCR only allows the study of one gene at a time, which makes the process very slow. The development of a new biotechnology, microarrays, has had a major impact on research into gene expression because it allows scientists to study thousands of genes all at the same time – giving the ability to study a genetic profile. Using a microarray we can identify which specific genes a cell is using at a particular point in time. This means that we can compare which genes are turned on or off in different conditions (e.g. when cancer is present compared to when cancer is absent).